Case Study: Bringing the Power of HPC to Drug Discovery and the Delivery of ‘Smarter’ Healthcare
In medieval times, leeches were used to "cure" arthritis and a number of other ailments. Today, while these rather repulsive little creatures are making a medical comeback of sorts, most people would rather treat creaky joints with less Draconian methods. Fortunately, there are companies like GNS Healthcare engaged in research to find new drug therapies to treat arthritis and many other common ailments. And their work is becoming even more critical as the United States and many other parts of the world undergo a rapid demographic shift. The baby boomers have reached their "golden years."
As Colin Hill, CEO and president of GNS, explains, "Rheumatoid arthritis is one of the primary diseases of old age, along with cancer, Alzheimer's and other infirmities. More than two million people in the US have rheumatoid arthritis, tens of millions more have other forms of arthritis, and even more will be afflicted as the population gets older. This represents a multibillion-dollar market for the drug industry from the sale of anti-inflammatory medications — such as anti-TNF therapies — which provide patients with great relief and hope. However, there is a problem. The National Institute of Health estimates that 20-30 percent of patients do not respond sufficiently to a given anti-TNF drug. Developing effective drugs that will benefit this population is a major research opportunity."
GNS, a privately-held biotechnology company headquartered in Cambridge, Mass., combines supercomputing with breakthroughs in genomics to put the power of modern mathematics and computation at the center of the drug discovery and development process. Specifically, the nine-year-old company was created to leverage data becoming available in the wake of the Human Genome Project and the latest advances in supercomputers to create data-driven models of human disease progression and drug response. These models are then simulated to discover novel drug targets, a critical step in creating new drug programs for diseases such as cancer, diabetes and, of course, rheumatism. By identifying genetic markers of response, GNS and its partners can tailor medicines to subpopulations or individual patients who are more likely to respond. This is a step toward personalized medicine in which a profile of a patient's genetic make-up can guide the selection of drugs or treatment protocols to ensure a more successful outcome and minimize side effects.
As recently as eight years ago, the application of high performance computing (HPC) techniques to drug discovery efforts was problematic at best. Using the best artificial intelligence platforms available at the time, even clusters composed of 40 or 50 processors could take up to 12 months to run through the DNA sequence data and corresponding gene expression and clinical response data needed to identify the important genes in a tumor when compared to normal tissue. Today, due to advances in supercomputing and software platforms from companies like GNS with its REFS computational environment, Wolfram Research with Mathematica, and The MathWorks with MATLAB, results of this type can now be achieved in weeks — and, Hill adds, "much more comprehensively."
REFS Data-Driven Process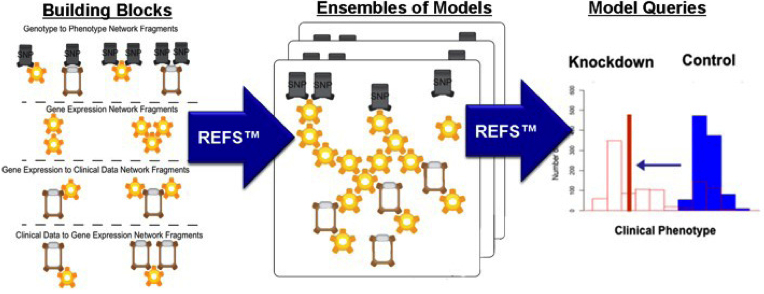
Figure 1: The REFS process begins with the creation of model building blocks, proceeds through the construction of an ensemble of models from the building blocks, and results in the simulation of the ensemble of models to extract quantitative outcome predictions accompanied by confidence levels.
Managing the Data Deluge with HPC and REFS
Only a supercomputer or high performance cluster can hope to churn through this deluge of data. "Today's advanced computational capabilities are essential, given the fact that the scale and complexity of the data derived from DNA sequencing machines and other sources has progressed tremendously in the last five to ten years," Hill says. "We have this strong conviction that the major game-changing advances in the biomedical sciences and drug development will not occur on a short time-scale without the extreme use of supercomputing."
GNS has developed a unique software platform that it calls "reverse engineering/forward simulation" (or REFS) that systematically turns multiple layers of disparate data types into an unprecedented view of a system of interest, rapidly performing billions of calculations to determine how the variables describing the system interact with and causally influence one another. These computer-assembled models are then queried rapidly through billions of in silico experiments to discover the most important genes and proteins driving the system's behavior, and to predict the system's behavior under previously unobserved conditions. (See Figure 1 for a graphical representation of REFS.)
For example, GNS recently leveraged the latest in HPC in a collaboration with Biogen Idec, one of GNS's key partners. The goal, as alluded to earlier, was to find relief for arthritis patients who are not responding to anti-TNF therapy. Thomas Neyarapally, senior vice president of corporate development for GNS, says, "Our mandate was to identify novel drug targets that would allow Biogen Idec to develop new, effective drugs for this patient population."
Not an easy task. The project involved gathering clinical data from 70 patients, including analyzing their DNA to look for single nucleotide polymorphisms (SNPs), which are DNA sequence variations in which a single nucleotide (A, T, C or G) in the genome sequence is altered. Also included were gene expression measurements from the blood. Clinical outcomes — such as each patient's number of swollen joints, pain levels and other kinds of blood markers — were also added to the mountain of data included in the study. The idea was to use GNS's supercomputer-driven capabilities to build models directly from these data. GNS could then use the models to conduct simulations that would indicate the best drug intervention points for individual patients based on their specific DNA and clinical background (see Figure 2).
REFS BioModels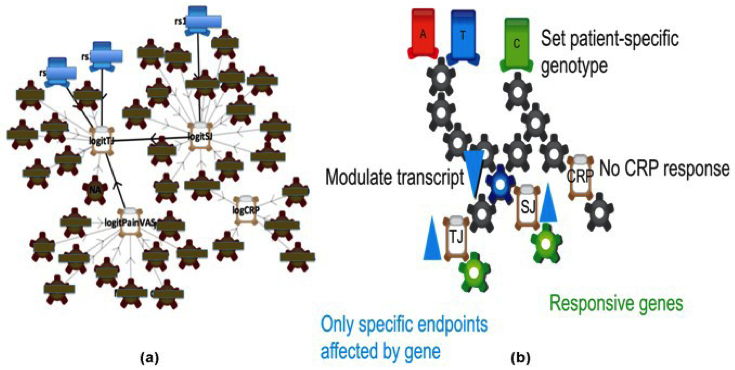
Figure 2: (a) A REFS model for the Biogen Idec project, compressed to show only links common to 30 percent or more of the 1000 networks. (b) A schematic showing how individual patient outcomes can be predicted using the REFS model. TJ = Tender Joints; SJ = Swollen Joints; CRP = C-Reactive Protein.
Before the model-building process begins, tens of thousands of variables that have relevant information content are identified. Then, the first computational step in this process is to evaluate trillions of mathematical relationships between these tens of thousands of variables and score the likelihood of the relationships based on the patient data. A large subset of potential relationships whose existence was highly improbable given the patient data were eliminated.
Even with this reduced dataset, the GNS platform had to explore how the robust relationships — analogous to puzzle pieces — fit together. Neyarapally says that for the Biogen Idec project, they started with trillions of puzzle pieces (or "network fragments") to create global computer models of drug responses connecting the thousands of variables. This means using the supercomputers to create and test not just a few models, but trillions of models which are scored against the patient data, with the puzzle pieces being swapped in and out to improve the model scores in an automated, iterative fashion. After the best scoring models have been created, these models are then used together as an ensemble of models; the ensemble is queried to generate a prediction of the best drug targets for the individual patient.
"A project of this kind involves hundreds of thousands of data points reflecting the individual points of genetic variation from patient to patient," Neyarapally explains. "Let's say one has a SNP chip that measures 500,000 points of genetic variation — that's 500,000 variables right there. Then one has the activity of genes being measured from the blood, adding another 20,000 to 40,000 measurements to the mix. And finally one has the clinical data — measurements describing tender and swollen joints, degree of pain, etc. — that provide another handful of variables."
"We deal with the uncertainty that's inherent in the data by using the ensemble of a thousand models as opposed to just one model," Neyarapally says. "By using supercomputers, we don't get just one answer to a given question — we're getting a thousand answers. And we are able to ask more than one question at a time. In fact, we can automatically ask millions of questions very rapidly." For example, REFS can generate a simulated "knock-down" of each gene (that is, a reduction in the activity of a gene), and then evaluate the impact of this knock-down on outcomes.
In a wet lab, the investigators would have to use a biological substance to reduce the activity of a particular gene, which over all the desired genes to test would take months and millions of dollars. Further, such experimentation cannot be done directly in humans. "Doing all this in silico," he says, "is significantly faster and allows us to look at many more gene interactions."
In addition, GNS's REFS platform is proving useful in other domains beyond the biological sciences. Indeed, the company just completed the venture capital financing of a separate subsidiary named Fina Technologies, Inc., that is focused on applying the REFS platform and other machine-learning tools to large-data problems in the worlds of financial trading, e-commerce, risk and advanced military intelligence.
"Essentially, what we are doing is automating aspects of the scientific method," Neyarapally continues. "Using supercomputers and our advanced software platforms, researchers can create hypotheses, rapidly test them and obtain answers with quantified confidence levels. Then we can either test these predictions in the lab or against an independent data set."
HPC: Allowing Otherwise Impossible Insights
Hill comments that — with respect to their core biological work — HPC allows GNS's scientists to perform experiments computationally that would be impossible to conduct on human subjects. "Unlike the design for an airplane engine or a computer circuit, organic systems such as people, the stock market or global weather do not come with a blueprint — a fact that makes conducting these kinds of experiments more difficult than the early proponents of computational biology and drug discovery expected. All the steps in the process are essential. Because these investigations are not engineering projects, one can't just jump to the final step and create a model that is accurate and has strong predictive power. First, one has to use the supercomputer and the multi-layered data to elucidate the building blocks — the puzzle pieces. Then, one uses the supercomputer to assemble those puzzle pieces by exploring many billions of configurations and scoring them. Then, one can move to the final step of creating the ensemble of models and doing the in silico experiments — the 'what if' hypothesis testing that is essential to validate the results."
Despite how integral supercomputers are to GNS's work, the company has only modest in-house HPC capabilities. More often GNS relies on the latest computational resources available from supercomputing vendors and its partners located around the world. "We buy time commercially from a number of sources," Hill says. "We use a combination of IBM Blue Gene and other machines — they're in the tens of thousands of processors. And we're essentially running on these machines every single day, every single week of the year. For us, it's just as easy to access supercomputing resources remotely as it is to work with in-house systems — maybe easier. The Internet has made it all possible."
He notes that in the biotech world, the stakes are very high. The drug development universe is unforgiving and binary — one either discovers a new drug molecule first or one's competitors do. It is winner take all — being second means losing out on the economic return from a drug that could generate billions of dollars a year.
"In our world, it's true that HPC has made GNS and our partners more competitive and profitable," Hill concludes. "And yes, it certainly has sped up our projects and dramatically cut the amount of time it takes to get answers. But I think one of the biggest benefits of HPC is that, inevitably, when scientists have more and more computing power in their hands, they ask bigger questions and tackle more ambitious projects. They are able to analyze far greater quantities of data. That's exactly what's been happening at GNS, allowing us to provide cutting-edge answers to partners like Pfizer and Biogen Idec in rheumatoid arthritis, and Johnson & Johnson in cancer. We are now able to systematically evaluate trillions of building blocks and end up with very powerful answers."
About the Project
This case study was produced as part of a project that was created to demonstrate the business and competitive value of modeling, simulation and analysis with HPC in the U.S. private sector, motivate usage of this innovation-accelerating technology throughout the DoD's supply chain, and identify technologies and partners that can help support an HPC infrastructure for the DoD supply chain base. It was led by the University of Southern California's Information Sciences Institute (ISI) and supported by funding from the Defense Advanced Research Projects Agency (DARPA) under contract number FA8750-08-C-0184. DARPA is the central research and development office for the U.S. Department of Defense. DARPA's mission is to maintain the technological superiority of the U.S. military and prevent technological surprise from harming our national security. For more information, see http://www.darpa.mil.
Complete information about the other case studies and pilot programs associated with this project is available at http://www.compete.org/hpc/darpapilots/.
Reprinted with permission of the Council on Competitiveness










